Caris GPSai
Identify Tumor Origin and Improve Treatment Decisions with Caris GPSai
Cancer of Unknown Primary (CUP) diagnosis is typically treated empirically and has very poor outcomes, with median overall survival less than one year.1 Caris GPSai™, a Genomic Probability Score, uses whole exome (DNA) and whole transcriptome (RNA) sequencing coupled with trained and validated neural networks to aid in identifying the tissue of origin.
Caris GPSai analyzes genomic and transcriptomic data to match a tumor’s molecular signature to one or more of 90 cancer categories from the Caris database. Caris GPSai provides additional insight to help oncologists better manage CUP or cases with atypical clinical presentation or clinical ambiguity.
Caris GPSai™ is included for all CUP cases and also may be selected on the requisition for other tumor types.
Enhance Diagnostic Confidence with Artificial Intelligence
Considering that rates of inaccurate diagnosis range between 3% and 9%2, Caris GPSai provides an integral part of quality control and may lead to improved diagnostic accuracy.
The Caris GPSai algorithm was trained and retrospectively validated on over 250,000 cases from the Caris database. The algorithm was then validated prospectively on an additional 3,200 MI Tumor Seek Hybrid™ cases.
VALIDATION STUDY RESULTS

Accuracy
Validation based on non-CUP top 1 calls across 90 tumor categories in 3,200 MI Tumor Seek Hybrid cases
Sample Output3 For Cholangiocarcinoma (95%)
TISSUE
-
Adrenal Cortical Carcinoma (score 0.00)
-
Bladder/Urinary Tract (score 0.00)
-
Bowel (score 0.00)
-
Breast (score 0.00)
-
CNS/Brain (score 0.00)
-
Cervix/Uterine Carcinoma (score 0.00)
-
Cutaneous Squamous Cell Carcinoma (score 0.00)
-
Esophagus/Stomach (score 0.00)
-
Germ Cell Tumor (score 0.00)
-
Hematological (score 0.00)
-
Hepatocellular Carcinoma (score 0.03)
-
Kidney (score 0.00)
-
Melanoma (score 0.00)
-
Mesothelioma (score 0.00)
-
Neuroendocrine Neoplasm (score 0.00)
-
Non-Small Cell Lung Carcinoma (score 0.00)
-
Orogenital Squamous Cell Carcinoma (score 0.00)
-
Ovarian Epithelial Tumor (score 0.00)
-
Pancreatobiliary (score 0.97)
-
Peripheral Nervous System (score 0.00)
-
Prostate Adenocarcinoma (score 0.00)
-
Salivary Gland Tumor (score 0.00)
-
Sex Cord Stromal Tumor (score 0.00)
-
Soft Tissue/Bone (score 0.00)
-
Thymic Carcinoma (score 0.00)
-
Thyroid
Pancreatobiliary Expanded
-
Cholangiocarcinoma (score 0.95)
-
Gallbladder Cancer (score 0.00)
-
Pancreatic Adenocarcinoma (score 0.02)
Caris GPSai Tumor Origin Predictor
Adrenal Cortical Carcinoma
Bladder/Urinary Tract
- Urothelial Carcinoma
- Bladder Adenocarcinoma
Bowel
- Appendiceal Adenocarcinoma
- Colorectal Adenocarcinoma
- Small Bowel Carcinoma
Breast
- Metaplastic Breast Cancer
- Breast Invasive Lobular Carcinoma
- Breast Invasive Ductal Carcinoma
CNS/Brain
- Diffuse Glioma
- Meningioma
Cervix/Uterine Carcinoma
- Uterine Serous Carcinoma/Uterine Papillary Serous Carcinoma
- Uterine Carcinosarcoma/Uterine Malignant Mixed Mullerian Tumor
- Uterine Clear Cell Carcinoma
- Cervical Adenocarcinoma
- Uterine Endometrioid Carcinoma
Cutaneous Squamous Cell Carcinoma
Esophagus/Stomach
- Esophageal Squamous Cell Carcinoma
- Esophagogastric Adenocarcinoma
- Stomach Adenocarcinoma
- Adenocarcinoma of the Gastroesophegeal Junction
- Esophageal Adenocarcinoma
Germ Cell Tumor
Hematological
Hepatocellular Carcinoma
Kidney
- Renal Cell Carcinoma
- Chromophobe Renal Cell Carcinoma
- Papillary Renal Cell Carcinoma
- Renal Clear Cell Carcinoma
- Wilms Tumor
Melanoma
Mesothelioma
Neuroendocrine Neoplasm
- Well/Moderately-Differentiated Neuroendocrine Tumor
- Poorly-Differentiated Neuroendocrine Carcinoma
- Large Cell Neuroendocrine Carcinoma
- Small Cell Neuroendocrine Carcinoma
- Paraganglioma/Pheochromocytoma
- Merkel Cell Carcinoma
Non-Small Cell Lung Carcinoma
- Lung Squamous Cell Carcinoma
- Lung Adenocarcinoma
Orogenital Squamous Cell Carcinoma
Ovarian Epithelial Tumor
- Ovarian Carcinosarcoma/Malignant Mixed Mesodermal Tumor
- Serous Ovarian/Fallopian Tube/Peritoneal
- Low-Grade Serous Ovarian/Fallopian Tube/Peritoneal Cancer
- High-Grade Serous Ovarian/Fallopian Tube/Peritoneal Cancer
- Mocinous Ovarian Cancer
- Clear Cell Ovarian Cancer
- Endometrioid Ovarian Cancer
Pancreatobiliary
- Gallbladder Cancer
- Pancreatic Adenocarcinoma
- Cholangiocarcinoma
Peripheral Nervous System
- Schwannoma
- Neuroblastoma
- Malignant Peripheral Nerve Sheath Tumor
Prostate Adenocarcinoma
Salivary Gland Tumor
Sex Cord Stromal Tumor
- Granulosa Cell Tumor
- Sertoli-Leydig Cell Tumor
Soft Tissue/Bone
- Gastrointestinal Stromal Tumor
- Leiomyosarcoma
- Angiosarcoma
- Rhabdomyosarcoma
- Synovial Sarcoma
- Osteosarcoma
- Liposarcoma
- Chondrosarcoma
- Endometrial Stromal Sarcoma
- Ewing Sarcoma
Thymic Carcinoma
Thyroid
- Anaplastic Thyroid Cancer
- Well-Differentiated Thyroid Cancer
- Papillary Thyroid Cancer
- Follicular Thyroid Cancer
- Hurthle Cell Thyroid Cancer
- Medullary Thyroid Cancer
For CH, EEA, EU and UK countries, Caris GPSai reports results for 21 cancer types.
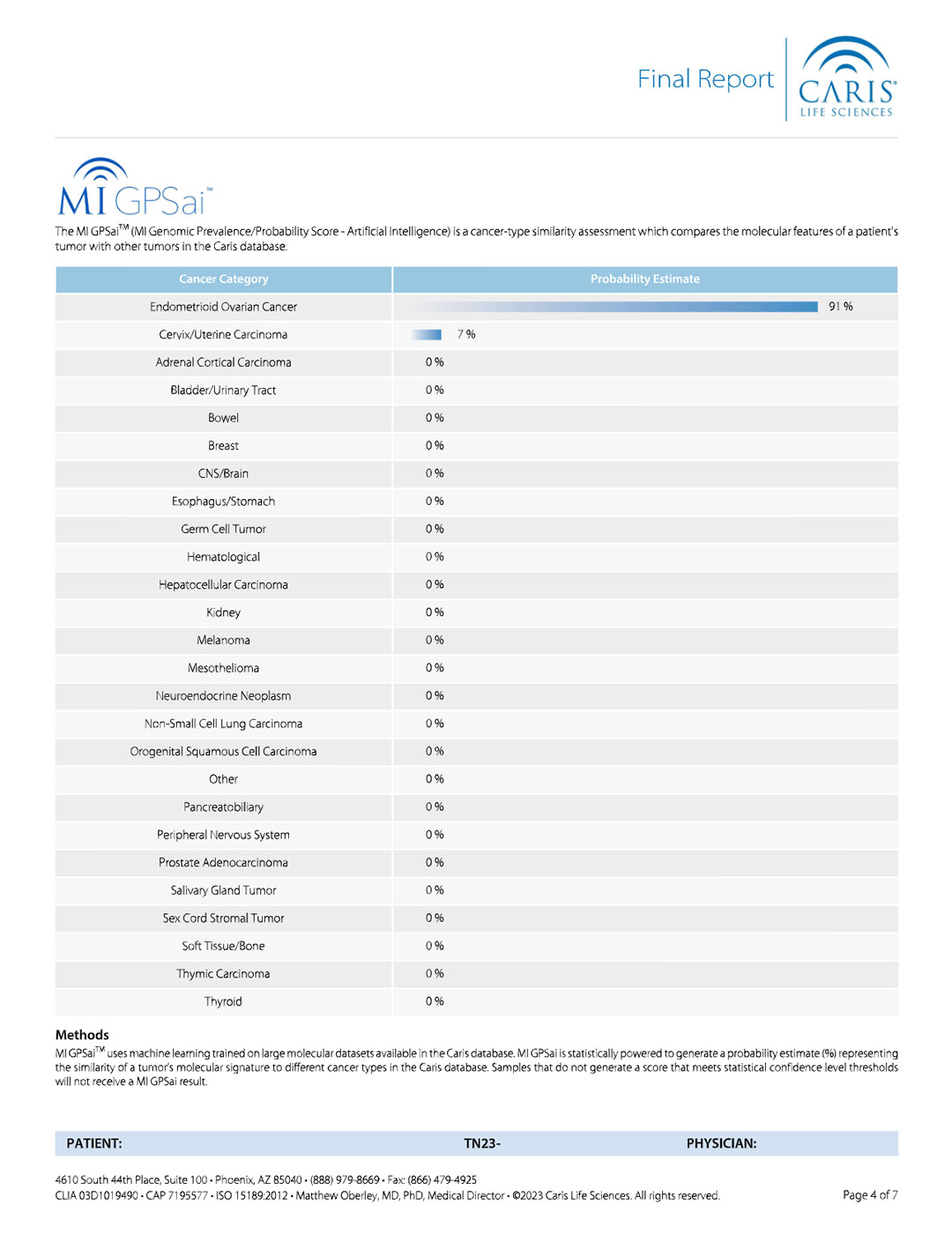
Example Caris Report: Caris GPSai (Genomic Probability Score)
- Reports a probability score based off both genomic (DNA) and now transcriptomic (RNA) profiling data.
- Analyzes genomic and transcriptomic data to match a tumor’s molecular signature to one or more of 90 cancer categories from the Caris database.
- Can be added to any solid tumor order at the oncologist’s discretion by selecting the appropriate box on the requisition. The Intent is for the test to only be requested for CUP and cases with atypical clinical presentation or clinical ambiguity.
- Must be requested by selecting the appropriate box on the requisition.
For CH, EEA, EU and UK countries, Caris GPSai reports results for 21 cancer types.
- Massard C, Loriot Y, Fizazi K. Carcinomas of an unknown primary origin–diagnosis and treatment. Nat Rev Clin Oncol. 2011 Nov 1;8(12):701-10. doi: 10.1038/nrclinonc.2011.158. PMID: 22048624
- Peck, M., Moffat, D., Latham, B., Badrick, T., Review of diagnostic error in anatomical pathology and the role and value of second opinions in error preventions, J. Clin. Pathol. 71 (11) (2018) 995-1000.
- Graphic generated using OncoTree framework. See Kundra, R., et al (2021) OncoTree: A Cancer Classification System for Precision Oncology. JCO Clin Cancer Inform 5, 221-230